Molecular Cytogenetics
Molecular cytogenetics involves the combination of molecular biology and cytogenetics. In general this involves the use of a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which we use DNA probes labeled with different colored fluorescent tags to visualize one or more specific regions of the genome. FISH can either performed as a direct approach, in which we use either metaphase chromosomes or interphase nuclei, or as an indirect approach, which involves the use of comparative genomic hybridization (CGH) analysis to asses the entire genome for of imbalanced chromosomal material.
Metaphase FISH
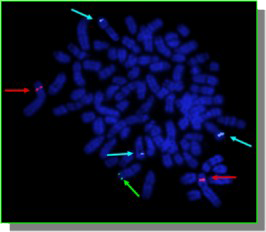
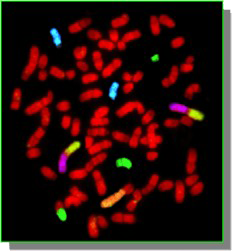
Metaphase and interphase FISH are “direct” approaches that make use of chromosome-specific paints or single locus probes. These are powerful techniques that allow a rapid assessment of the gross numerical and structural characteristics of a cell, looking directly at the chromosomes of the tumor cells Tumor metaphase FISH requires the availability of viable cells from which to produce tumor chromosome preparations for analysis. Interphase FISH is able to make use of preserved cellular material and may thus be extended to include the use of paraffin embedded biopsy sections. In the examples below, the figure on the left shows the application of FISH of single locus probes (SLP) to determine the number of copies of three different chromosomes in a canine cancer metaphase spread. In this case the chromosome with a blue SLP is present as three copies and so the cell is trisomy for this chromosome. On the right hand side is an example of a five color FISH reaction, using chromosome paint probes, to identify the nature of multiple aberrant chromosome in a metaphase preparation of a canine tumor cell.
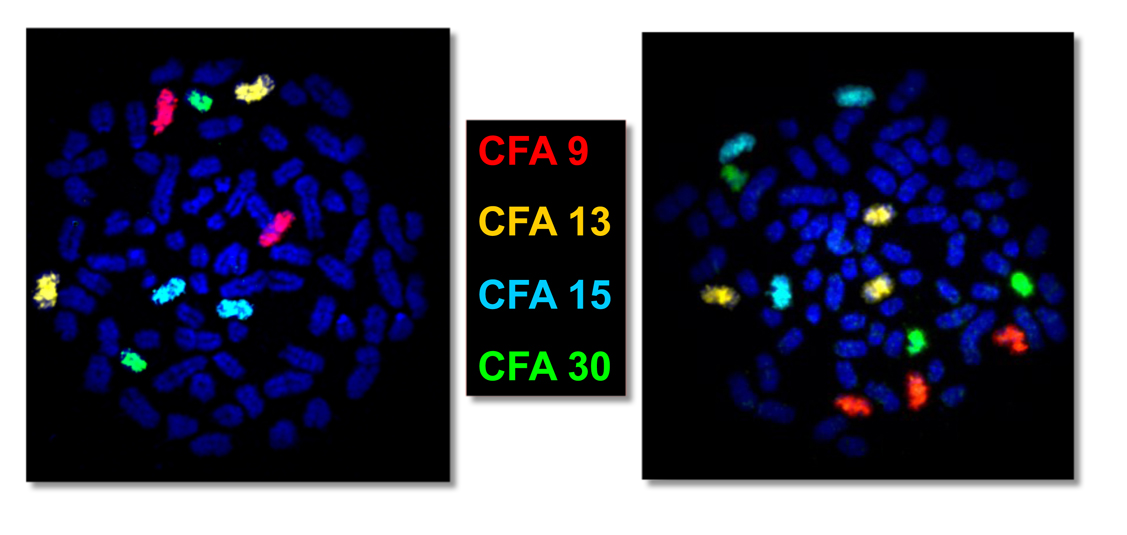
The use of whole chromosome paint probes allows us to determine the identity and copy number of extra chromosomes present in cancer cells. In the example below, both metaphase prepaprations have been probed with whole chromosome paint probes for dog chromosome (CFA) 9, 13, 15 and 30, using four different fluors (red, gold, aqua and green) as indicated. On the left, it is evident that there are two copies of all four chromosomes being evlauted. On the right each of these targeted chromosomes is present as three copies (trisomy) in this cell.
Whole chromosome paint probes are useful for looking at whole chromosome changes, but they are not able to reveal the presence of intrachromosomal structural changes. For this purpose we have assembled a panel of almost 2,500 canine BAC clones, each integrated into the canine genome assembly with approx. 1Mb intervals.Each clone has been shown to have a unique cytogenetic location when used in FISH and the genome wide panel has been organized into chromosome specific sets for all 38 autosomes and the X chromosome, at both 1Mb and 10Mb intervals. The example below shows the co-hybridization of 15 BAC clones labeled in five colors; seven map to CFA 9 and eight map to CFA 10, both at approximately 10Mb intervals. On the left is the whole metaphase image showing the chromosomes counterstained in DAPI and the cytogenetic location of all 15 BACs. In the middle is a schematic of CFA 9 and 10 showing the expected location of the BACs used, along with their corresponding colors (arrowed). On the rght the CFA 9 and CFA 10 homologues from the whole metaphase presented are shown, correclty oriented, demonstrating the spatial spearation of the probes along the lenght of eeach chromosome.
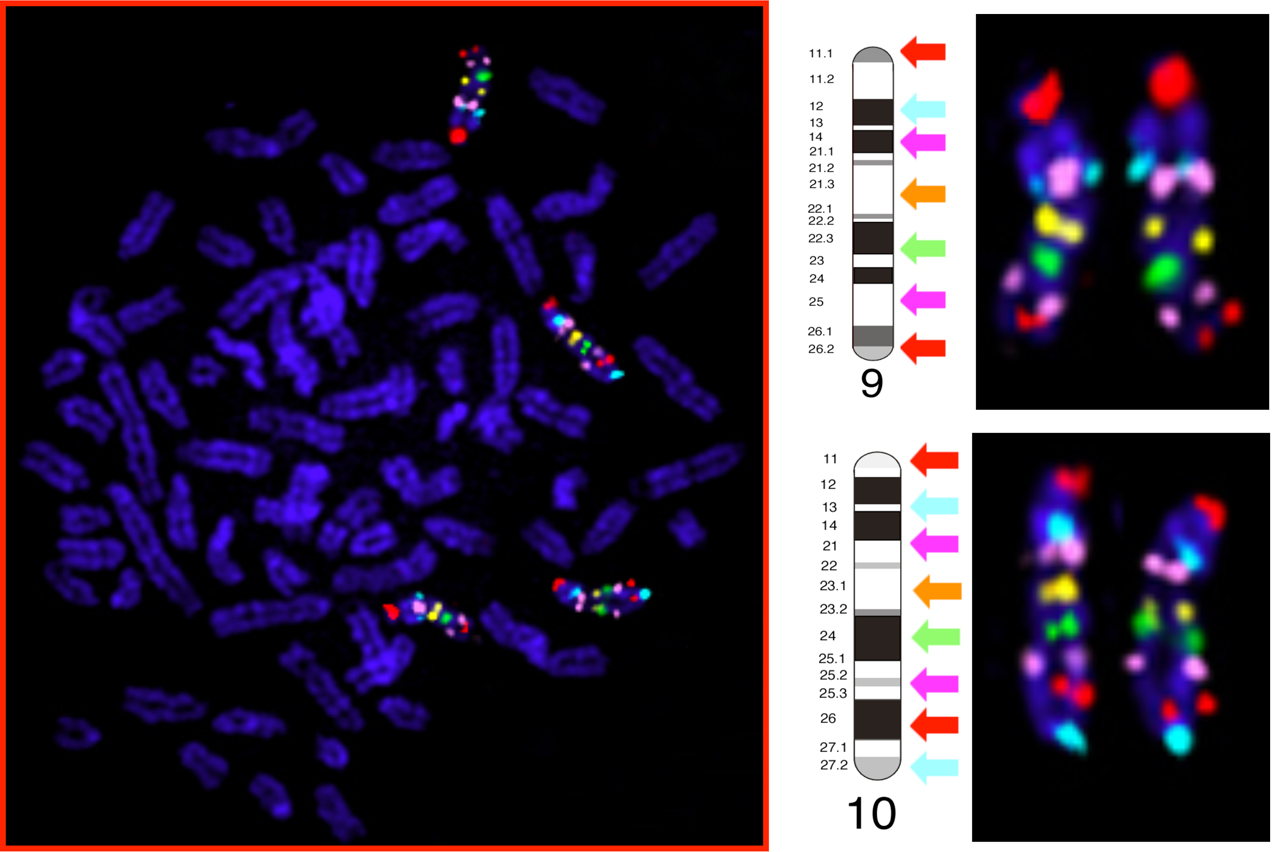
Using these kinds of tools, the resulting multicolor bar code developed for each chromosome allows us to identify where chromosome have undergone intrachromosomal structural changes, such as inversions, deletions and insertions.
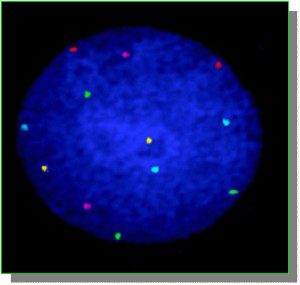
Interphase FISH
As with metaphase FISH, interphase FISH is a ‘direct’ approach where single locus probes (SLPs) are use to probe cell nuclei to assess the gross numerical and structural characteristics of a tumor cell population. In contrast to metaphase FISH interphase FISH is able to make use of preserved cellular material and may thus be extended to include the use of archival paraffin embedded biopsy sections. The example below shows the application of multicolor FISH of single locus probes (SLP) to determine the number of copies of five different chromosomes in a canine cancer cell nucleus. In this case the chromosomes represented by red, orange and pink SLPs all have a normal copy number of n=2, while the chromosomes represented by a blue and a green SLP are both present as three copies, indicating that both these chromosome are trisomy in this cell.